iyeung144.github.io
My Github Pages
Fitting Data: Hong Kong Air Pollutants
by Yeung Ka Ming, CFA
Summary
Fitting Data: Hong Kong Air Pollutants
R Libraries
library(fitdistrplus)
Preparation of data
airpollutants <- read.csv("airpollutants.csv", header = TRUE, sep = ",", encoding ="UTF-8")
head(airpollutants)
## StationName DateTime NO2 O3 SO2 CO PM10 PM2.5
## 1 Central/Western Thu, 24 Feb 2022 22:00:00 +0800 79.7 35.3 4.5 - 35.7 25.5
## 2 Central/Western Thu, 24 Feb 2022 23:00:00 +0800 98.1 19.1 4.4 - 37.2 25.9
## 3 Central/Western Fri, 25 Feb 2022 00:00:00 +0800 79.0 29.5 3.6 - 35.8 24.9
## 4 Central/Western Fri, 25 Feb 2022 01:00:00 +0800 72.6 29.1 2.7 - 32.4 22.7
## 5 Central/Western Fri, 25 Feb 2022 02:00:00 +0800 36.2 58.9 2.3 - 27.3 20.6
## 6 Central/Western Fri, 25 Feb 2022 03:00:00 +0800 32.5 50.4 2.3 - 24.4 19.2
fitdistrplus accepts vector only[1]
airdata <- as.numeric(airpollutants$NO2) # change to numeric but introduced NAs
airdata <- c(na.omit(airdata)) # omit NAs and covert data to a vector. fitdistrplus accepts numeric vector only
Plot data
plot(airdata,pch=20)
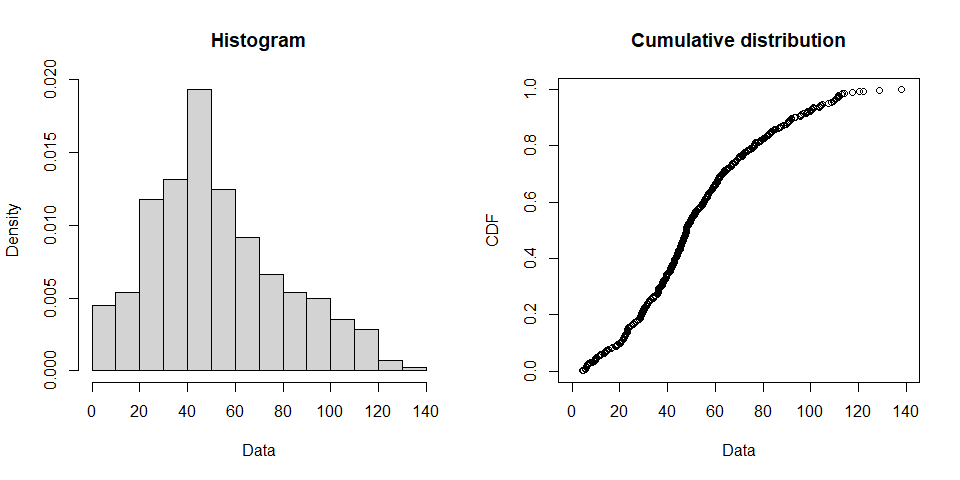
plotdist(airdata, histo = TRUE, demp = FALSE)
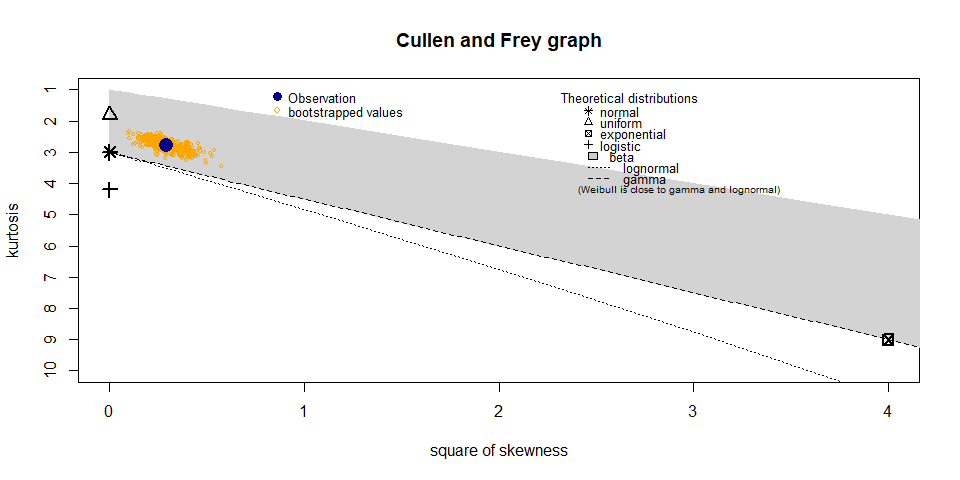
descdist(airdata,discrete = FALSE,boot = 500)
## summary statistics
## ------
## min: 4.3 max: 137.9
## median: 47.9
## mean: 52.55694
## estimated sd: 27.4781
## estimated skewness: 0.5394571
## estimated kurtosis: 2.780306
Fitting data
fit_w <- fitdist(airdata,"weibull")
fit_g <- fitdist(airdata, "gamma")
fit_ln <- fitdist(airdata, "lnorm")
summary(fit_w)
## Fitting of the distribution ' weibull ' by maximum likelihood
## Parameters :
## estimate Std. Error
## shape 1.997389 0.07620992
## scale 59.273086 1.51460174
## Loglikelihood: -1989.855 AIC: 3983.71 BIC: 3991.814
## Correlation matrix:
## shape scale
## shape 1.000000 0.311239
## scale 0.311239 1.000000
summary(fit_g)
## Fitting of the distribution ' gamma ' by maximum likelihood
## Parameters :
## estimate Std. Error
## shape 3.06166970 0.199430160
## rate 0.05824908 0.004121638
## Loglikelihood: -1998.918 AIC: 4001.836 BIC: 4009.94
## Correlation matrix:
## shape rate
## shape 1.0000000 0.9201068
## rate 0.9201068 1.0000000
summary(fit_ln)
## Fitting of the distribution ' lnorm ' by maximum likelihood
## Parameters :
## estimate Std. Error
## meanlog 3.789776 0.03160773
## sdlog 0.651610 0.02234980
## Loglikelihood: -2031.672 AIC: 4067.345 BIC: 4075.449
## Correlation matrix:
## meanlog sdlog
## meanlog 1 0
## sdlog 0 1
Compare different fitting results
par(mfrow=c(2,2))
plot.legend<-c("Weibull","gamma", "lognormal")
denscomp(list(fit_w,fit_g,fit_ln), legendtext=plot.legend)
cdfcomp(list(fit_w,fit_g,fit_ln), legendtext = plot.legend)
qqcomp(list(fit_w,fit_g,fit_ln), legendtext = plot.legend)
ppcomp(list(fit_w,fit_g,fit_ln), legendtext = plot.legend)
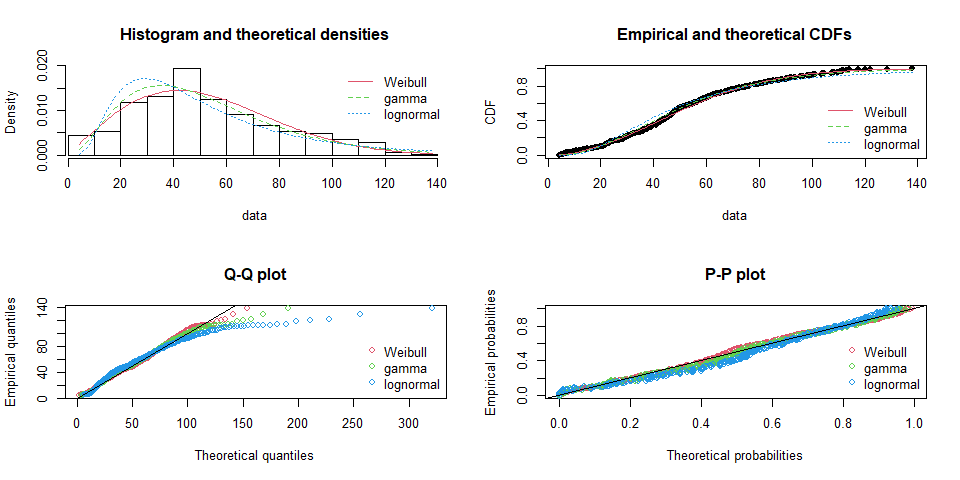
Goodness-of-fit test
gofstat(list(fit_w,fit_g,fit_ln), fitnames = c("weibull","gamma","lnorm"))
## Goodness-of-fit statistics
## weibull gamma lnorm
## Kolmogorov-Smirnov statistic 0.03711893 0.06390271 0.1052155
## Cramer-von Mises statistic 0.12721977 0.28117347 1.0806864
## Anderson-Darling statistic 0.86932594 1.99729639 7.0892961
##
## Goodness-of-fit criteria
## weibull gamma lnorm
## Akaike's Information Criterion 3983.710 4001.836 4067.345
## Bayesian Information Criterion 3991.814 4009.940 4075.449
Reference
[1] https://stackoverflow.com/questions/37091580/how-to-exclude-nas-fitdist-function
tags: fittingData